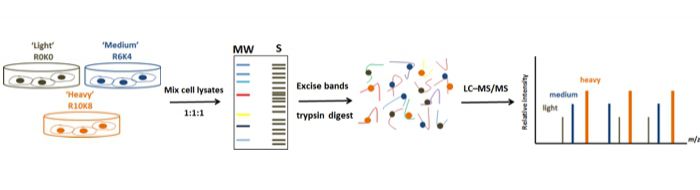
Stable isotope labeling with amino acids in cell culture (SILAC)-coupled MS analysis represents one of the most promising comparative quantitative methods that has been broadly employed in proteomic research generating vast amounts of functional data. This approach enables clear identification and quantification of protein dynamics essential in oncogenesis and therefore has been extensive utilized in cancer-proteome studies.
- PAPER: Deciphering the Tyrosine Kinase-regulated proteome in breast cancer (Molecular & Cellular Proteomics, 2015)
-
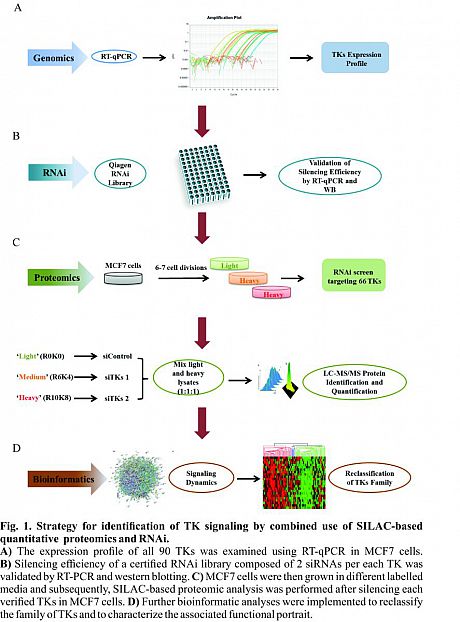 Recently, for the first time we described the global mapping of Tyrosine Kinases(TK)-regulated proteome using a high throughput RNAi screen combined with SILAC-based quantitative proteomics in MCF7 breast cancer cells (Fig. 1).
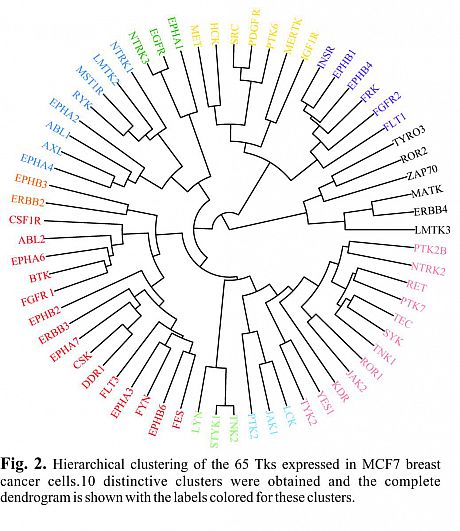
Recently, for the first time we described the global mapping of Tyrosine Kinases(TK)-regulated proteome using a high throughput RNAi screen combined with SILAC-based quantitative proteomics in MCF7 breast cancer cells (Fig. 1). Overall, four thousand distinct proteins were detected and quantified in the TK-silencing datasets showing a diverse landscape of modulated proteins. Based on the similarity in their proteomic changes, we presented 10 new distinctive clusters from the 65 TKs and ultimately characterized a unique proteomic signature and functional portrait of each cluster (Fig. 2).
These data suggest that despite the primary structural homology of kinases, their regulated proteome can vary significantly and may depend on other factors, notably their dynamic interactions with other proteins.
Our defined functional analysis of the TK-regulated proteome supports a fundamental involvement of TKs in the major cellular processes and highlights a compensatory activation in their associated signaling. In addition, our established database present a unique resource that can be utilized for comparative and integrative analyses, for inspection of function of each TK of interest, and their associated signaling networks in cancer.
- PAPER: Proteomic profile of KSR1-regulated signalling in response to genotoxic agents in breast cancer (BCRT, 2015)
-
Kinase suppressor of Ras 1 (KSR1) has been shown to contribute to tumorigenesis in multiple cancers, including skin, pancreatic and lung carcinomas.
However, our recent study suggested KSR1 as a tumour suppressor in breast cancer, the expression of which is potentially correlated with chemotherapy response.
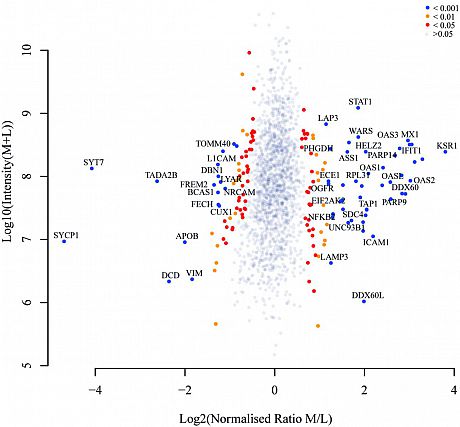
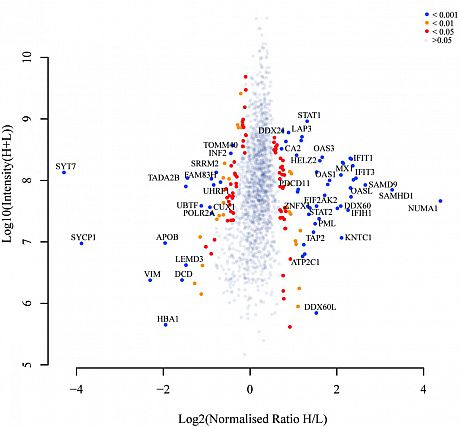
To further elucidate the KSR1-regulated signaling in response to genotoxic agents in breast cancer, we implemented SILAC-MS to globally compare cellular protein expression levels induced by KSR1 in the presence of doxorubicin (left volcano plot) or etoposide (rightvolcano plot).
Our data illustrated a comprehensive functional network that KSR1 is involved in and highlighted its significance in predicting chemotherapy response in patients.