Molecular mechanism(s) of chromosome segregation and genome rearrangements
Inheritance of accurate genetic material is crucial for cell renewal, functions, and continuity of life. Cells have evolved robust systems to precisely duplicate, repair, and transmit DNA that presumably prevent the transmission of genetic errors from generation to generation. However, the genetic makeup of (pre)cancerous cells becomes inexplicably altered in a cumulative manner. It is believed that the accumulation of genetic rearrangements drives the initiation and evolution of malignancy. Nevertheless, the origin(s) and mechanism(s) of early neoplastic transformation remain enigmatic.
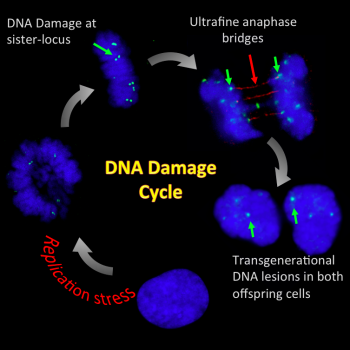
DNA damage cycle
To complete a cell division cycle, cells have to precisely duplicate and equally distribute their entire genomes into offspring cells. However, replication stress such as lack of sufficient nucleotides or inhibition of DNA polymerases can severely hinder replication processes and prevent its completion. As a result, replication stress can induce DNA damages and accumulation of recombination and/or replication intermediate structures. Strikingly, we found that this causes persistent sister-chromatid bridging (or the formation of so-called ultrafine DNA bridges) in mitosis, presumably due to unresolved recombination/replication structures. The improper disjunction of sister-chromatid bridges could result in the transmission of the damaged chromosomes to both daughter cells and threaten their genome integrity. The intertwining DNA structures are recognised by a number of DNA repair factors, defects of which result in human cancer-prone human disorders, Fanconi anemia and Bloom syndrome. Intriguingly, the resultant DNA lesions are persistent throughout the entirety of next G1 even though they are recognised by a number of DNA repair factors including 53BP1 protein. This suggests that the “transgenerational DNA lesions” might be constantly under surveillance between generations. It is conceivable that if replication stress is persistent, a DNA damage cycle is initiated and that could gradually and accumulatively rearrange genetic makeup leading to cancer formation.
Therefore, we are interested in understanding how cells achieve faithful chromosome segregation in normal and replication stress conditions, focusing on the areas of
- how replication stress-associated DNA structures hinder mitotic progress and sister chromatids disjunction;
- how misregulation of mitosis leads to chromosome breakage, particularly on common fragile sites (CFSs) and centromeres rupture;
- how the mitosis-induced DNA damage triggers gross chromosomal rearrangements, e.g. Robertsonian translocation and isochromosome formation, in offspring daughter cells;
- how the transgenerational DNA damage initiates cell transformation.
Our goal is to understand the molecular mechanism(s) of genome rearrangements and develop preventative medicine and targeting therapy for cancers.
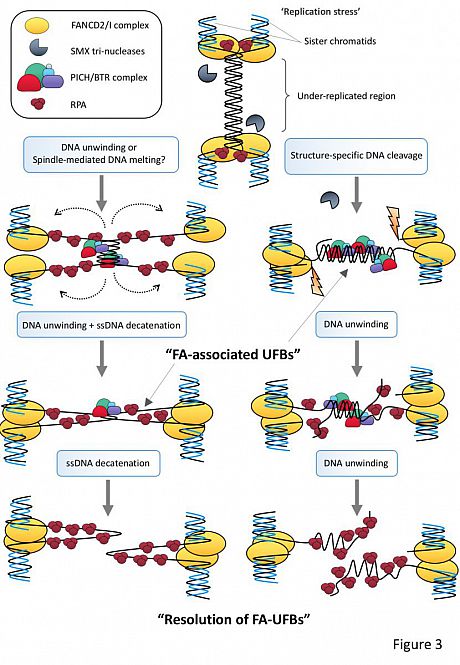
Below: Possible pathways to resolve under-replicating DNA regions during mitosis via either single-stranded DNA decatenation (left) or branched structure cleavage (right).