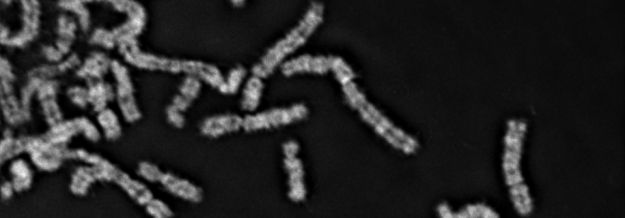Chromosome Dynamics and Stability Group
The preservation of genome integrity is of paramount importance for the continuity of life. This is achieved through a series of highly coordinated cellular processes including accurate DNA replication, repair, and faithful chromosome segregation. Our group is interested in the interplay between DNA replication and chromosome segregation, and in how cells equally distribute sister chromatids during mitosis. We study the mechanisms of sister chromatids cohesion, chromosome biorientation, and disjunction. Notably, we focus on how sister-DNA intertwining structures influence chromosome segregation and integrity. Accumulating evidence has shown that disturbance of these processes can lead to transgenerational DNA lesions. We thus also investigate how mitosis-associated DNA damage may initiate cancer genome evolution. By employing high/super-resolution microscopy, we study how different parts of human chromosomes change during mitosis under normal or replication stress conditions. Additionally, combining CRISPR genome-editing techniques and conditional chemical inhibition, we study how mitotic kinases including Cyclin-dependent kinases, Polo-like kinase 1 and Aurora B participate in chromosome protection, and the resolution of sister DNA intertwinements, also known as ultrafine DNA bridges (UFBs). We believe that the misregulation of chromosome maintenance pathways in mitosis is one of the driving forces to gross genome rearrangements, leading to cell transformation and some rare human genetic disorders.
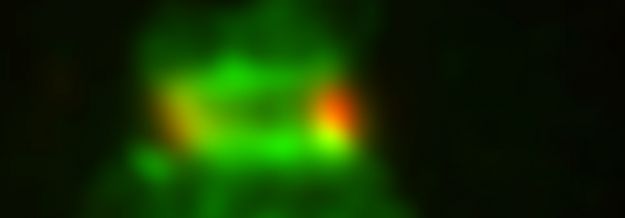
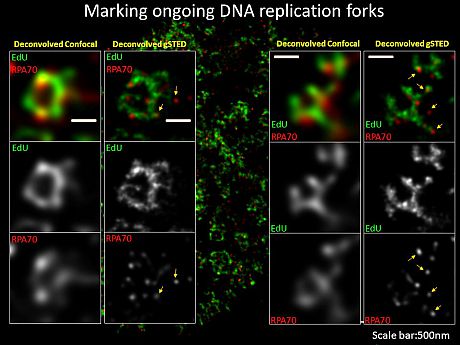
Below: employing Gated-STED super-resolution microscopy to locate components at ongoing DNA replication forks (co-labelled by EdU and RPA)
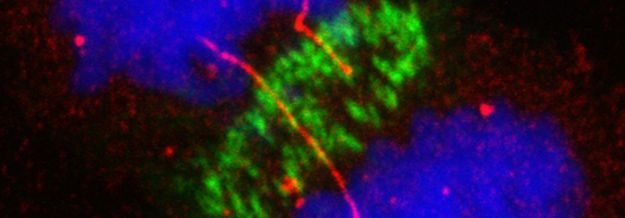
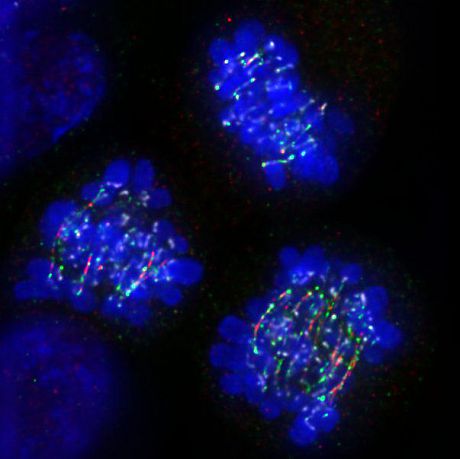
Below: HeLa anaphase cells expressing hypomorphic 53BP1 protein exhibit sister chromatids intertwining and rupture (red: gammaH2AX; green: PICH)
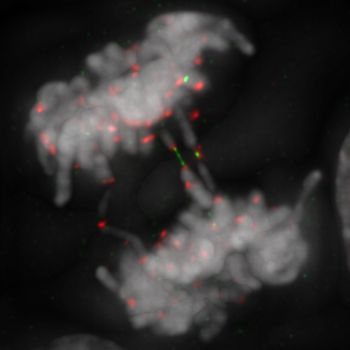
Below: Human RPE1 cells lose centromere integrity in early mitosis when Polo-like kinase 1 (PLK1) activity is compromised. This leads to severe breakages at centromeres, resulting in the formation of centromeric DNA threads coated by the ultrafine-DNA bridge binding complex and whole chromosome arms separation (Grey, centromeres; Red, RPA; Green, PICH translocase).
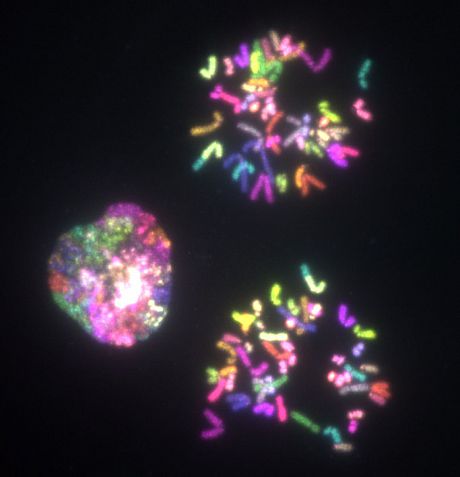
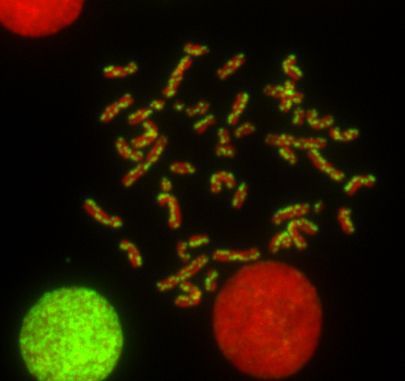
Below: Mitotic chromosomes of human diploid epithelium cells labelled with multiple-colour FISH probes for chromosome rearrangements analysis.
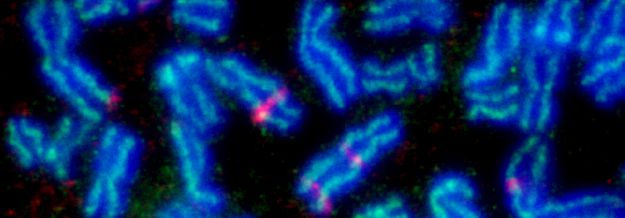
Below: Bloom's syndrome helicase (BLM) deficient cells show significant increases in sister chromatids exchange (SCE).
"If you are locating in low- or middle-income countries, you can apply the following access grants to obtain microscopy traininng from us."
https://globalbioimaging.org/i4a#who-can-apply
Our research is supported by