BRCT domains, so-called due to their identification at the C-terminus of the BRCA1 protein, act as protein-protein interaction modules. They frequently occur in pairs, but can exist as singletons or in threes (e.g. in TopBP1). BRCT domain interactions are either phospho-dependent or -independent. Phospho-specific interactions occur via a conserved phosphopeptide binding site found in proteins such as Mdc1, 53BP1 and Crb2, where generally one BRCT domain in a BRCT pair contains the binding site.
BRCT (BRCA1-C-terminal) domains, first identified in S. pombe Rad4/Cut5 (Fenech et al 1991) are involved in a range of protein-protein interactions, involving the BRCT domains as well as the inter-BTCT linker e.g. Watts & Brisset, 2010. As their name suggests they are present at the C-terminus of the BRCA1 protein, mutations in which are responsible for a high proportion of breast and ovarian cancers. They are also present in many other DNA damage response proteins, including 53BP1, a protein that interacts with the tumour suppressor protein p53.
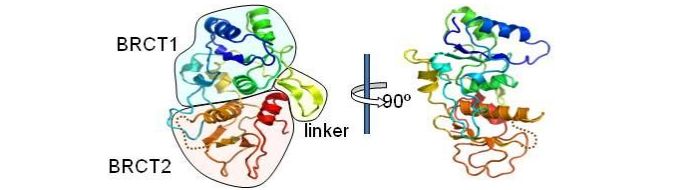
Using fission yeast as a model system, we initially investigated the role of the BRCT domains in the S. pombe homologue to 53BP1, Crb2, and the nature of the phosphorylation dependent interaction in which it is involved. Crb2 is a key player in the DNA integrity checkpoint process in S. pombe (Willson et al., 1997). In collaboration with Laurence Pearl's group, we elucidated the structure of the BRCT domains in Crb2 (Kilkenney et al., 2008). Using information gained from this structure we created and analysed two sets of 'separation of function' mutants. One class of mutants, mutated in the inter-BRCT linker region, was very sensitive to DNA damaging agents and defective in the DNA integrity checkpoint, while the other set was less sensitive to damage, and likely represented a defect in repair of damage.
Recently we have been focussing our attention on 53BP1, the human orthologue of Crb2. 53BP1 has a key role in determining repair pathway choice: it is thought to promote non-homologous recombination (NHEJ) and prevent homologous recombination. Following exposure to DNA damaging agents 53BP1 is rapidly recruited to nuclear foci. Initial recruitment is mediated by two post-translational modifications: via H2AK13/15-anchored ubiquitin chains and by direct interaction of the tandem Tudor domains of 53BP1 with dimethylated H4K20. We have shown that in addition to this, 53BP1 interacts with gamma H2AX, the primary mark of DNA damage (Baldock et al., 2015). This interaction is required for slow kinetics repair of DNA double strand breaks in the G1 phase of the cell cycle. This fraction of the repair events represents complex DNA breaks, and/or breaks present in heterochromatin. We have demonstrated that interaction of 53BP1 with gamma H2AX is required to retain ATM (ataxia telangiectasia mutated) at sites of DNA damage.
We have also collaborated with labs outside Sussex e.g. with the labs of Prof Martin Bushell (U of Leicester) and of Jo Morris (U of Birmingham). Results from these collaborations have demonstrated that recruitment of DNA repair factors, such as 53BP1, is dependent on Drosha (Lu et al., 2018), and that relocalisation of 53BP1 at extended times after DNA damage is dependent on the BRCA1-BARD1 ubiquitin ligase activity and SMARCAD1 (Densham et al., 2016).