For a general description of research in the West lab please click on the 'non-specialists' tab.
There are four main research themes in the West lab at present:
1. B cell reprogramming by EBV through 3D chromatin reorganisation (Blood Cancer UK)
2. The regulation and function of the cell-cycle regulator RGC-32 in EBV-infected cells (MRC)
3. The consequences of natural sequence variation on the structure and function of EBV nuclear antigens (MRC with Paul Farrell and Rob White, Imperial College)
4. Drug discovery using more widespread EBV variants (Wellcome Trust ISSF and Cancer Research UK)
1. B cell reprogramming by EBV through 3D chromatin reorganisation
Transcriptional reprogramming of B cells by EBV plays a central role in immortalisation, a critical step in lymphoma development. Understanding the mechanisms involved in B cell reprogramming by EBV is central for the development of new therapeutic strategies. All six of the nuclear proteins (Epstein-Barr nuclear antigens; EBNAs) expressed by Epstein-Barr virus in latently infected immortalised B-cells can function as transcriptional regulators. Our research primarily focuses on four key nuclear antigens; EBNA 2, EBNA 3A, EBNA 3B and EBNA 3C that play a central role in controlling both viral transcription and cellular gene transcription. Interestingly, none of these key EBV-encoded transcriptional regulators can bind DNA directly and interact with their viral and cellular response elements by hijacking cellular DNA-binding proteins such as RBP-Jkappa, PU.1 and EBF-1. Using chromatin immunoprecipitation-sequencing (ChIP-seq) we showed that EBNA 2 and EBNA 3 proteins primarily target long-range gene control elements to transcriptionally reprogramme cell genes (McClellan et al, 2012 and 2013) (Figure 1).
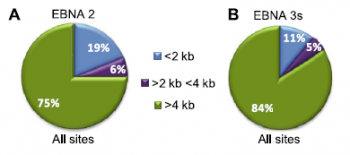 Figure 1. Analysis of ChIP-seq data for EBNA 2 and EBNA 3 proteins. (A) Pie chart showing the distribution of all significant binding sites for EBNA 2 relative to gene transcription start sites. (B) Distribution of EBNA 3 family binding sites.
Figure 1. Analysis of ChIP-seq data for EBNA 2 and EBNA 3 proteins. (A) Pie chart showing the distribution of all significant binding sites for EBNA 2 relative to gene transcription start sites. (B) Distribution of EBNA 3 family binding sites.
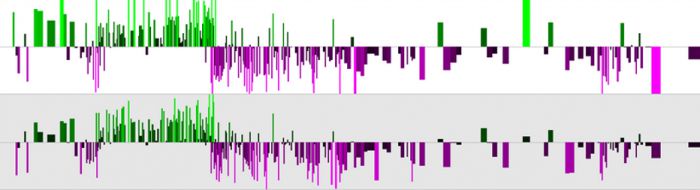
We have since studied a range of these long-range control elements using techniques including chromosome conformation capture (3C, 4C and capture Hi-C) to determine how they are used by EBNA 2 to activate genes and by EBNA 3 proteins to repress genes (Figure 2). Our work has shown how integrin and chemokine networks are targeted by EBV (McClellan et al, 2012), how enhancer looping can be manipulated in different ways by EBNA 3 proteins to repress genes (McClellan et al, 2013) and how the genes encoding RUNX transcription factors are controlled by EBV (Gunnell et al, 2016). Our work has also shown how MYC is activated by EBV through the reorganisation of a 3 Mb chromatin domain encompassing multiple super-enhancers (Wood et al, 2016) and how EBV induces expression of the oncogenic microRNA miR-155 (Wood et al, 2018).
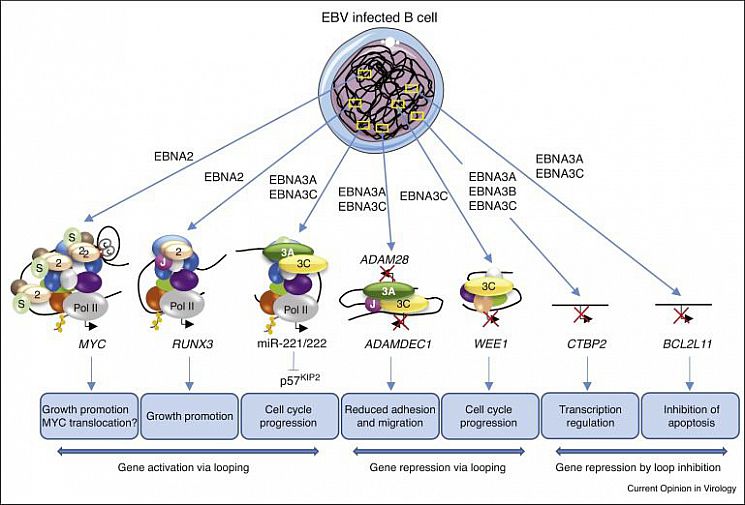 Figure 2. The effects of EBV transcription factors on enhancer–promoter interactions at different gene loci in B cells (miR-221/222 regulation was published by the Allday lab).
Figure 2. The effects of EBV transcription factors on enhancer–promoter interactions at different gene loci in B cells (miR-221/222 regulation was published by the Allday lab).
2. The regulation and function of the cell-cycle regulator RGC-32 in EBV-infected B-cells.
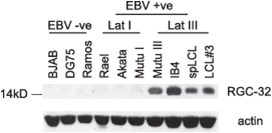 Figure 3. RGC-32 protein expression in B cell-lines.
Figure 3. RGC-32 protein expression in B cell-lines.
We discovered that protein expression of the cell-cycle regulator, Response Gene to Complement 32 (RGC-32 or C13ORF15), is upregulated in EBV-infected cells expressing the full panel of EBV latent genes (known as latency III) (Schlick et al, 2011) (Figure 3). EBV is known to disrupt cell cycle control through multiple mechanisms to promote B cell immortalisation. RGC-32 binds and activates the key mitotic kinase, CDK1, in a manner dependent on threonine 91 phosphorylation of RGC-32 by CDK1, indicating that RGC-32 may be a key regulator of G2/M progression. RGC-32 may therefore play a key role in EBV-mediated effects on mitotic entry. We are studying the mechanism of CDK1 activation by RGC-32 (with Jane Endicott, Newcastle), its role in mitosis (with Helfrid Hochegger, Sussex) and how it is regulated in EBV-infected cells. Our work has shown that RGC-32 is translationally repressed in EBV negative B cells through a mechanism that involves binding of the RNA-binding protein Pumilio to the RGC-32 3'UTR (Brocard et al, 2018). We are currently determining the further details of the mechanism of post-transcriptional control of RGC-32 (with Tracy Nissan, Sussex).
3. The consequences of natural sequence variation on the structure and function of EBV nuclear antigens
There is increasing evidence that natural variation in the EBV genome DNA sequence can affect the frequency or severity of disease, and affects normal EBV biology and infections. We are investigating two major aspects of EBV genome variation and their impact on the function of EBV nuclear antigens and the design of therapeutic strategies.
The largest natural variation of EBV divides virus strains into two types, called type 1 and type 2, which leads to striking differences in virus biology. They have different geographic distributions, with type 1 (which is more diverse) occuring worldwide, but type 2 having its highest incidence in sub-Saharan Africa and other parts of the world where other infectious diseases are very intense. Type 2 EBV is less effective at immortalising B cells in the laboratory and variation in the sequence of the EBNA 2 protein is known to be responsible for this. We have shown that sequence differences in type 2 EBNA2 create an additional binding site for a cellular repressor (BS69) that restricts the activation of cellular genes by type 2 EBNA2 (Ponnusamy et al, 2019) (Figure 4).
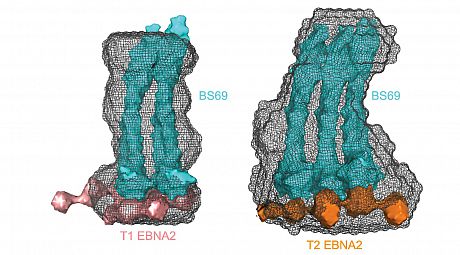 Figure 4. Small-angle X-ray scattering model showing that type 2 EBNA2 associates with an additional dimer of the cell repressor BS69.
Figure 4. Small-angle X-ray scattering model showing that type 2 EBNA2 associates with an additional dimer of the cell repressor BS69.
In collaboration with Paul Farrell and Rob White at Imperial College, London, we are further investigating how different types of EBNA 2 and EBNA 3 proteins work together in the transcriptional regulation of host cell genes.
We are also investigating the significance of diversity of the EBNA1 gene. EBNA1 is the only EBV protein made in all EBV associated cancers, and is essential to EBV persisting in those cancers, so it is an attractive target for therapy. The differences between EBNA1 protein sequences may affect its contribution to cancer and response to potential EBV drugs. We have identified the main variants of EBNA1 are investigating the functional consequence of variation in the EBNA1 DNA binding domain on its structure, DNA binding activity and senstivity to small molecule inhibitors. We are also studying the effect of sequence variation in EBNA1 on viral replication, EBV genome maintenance and interactions with protein partners in cell-based assays.
4. Drug discovery using more widespread EBV variants (Wellcome Trust ISSF and Cancer Research UK)
In new work we are exploring the use of EBNA1 variants in screens to identify small molecules that target proteins with sequences more representative of worldwide circulating strains.