Epstein Barr virus
Epstein–Barr virus is an almost ubiquitous human virus, which is transferred from person to person in saliva. Infection results in virus entry into both B-lymphocytes and epithelial cells. EBV promotes the proliferation of infected B-lymphocytes and readily generates immortalized cell lines when infection is undertaken in an in vitro culture system. The majority of these immortalized cells are recognized by the host immune system and destroyed but some enter the memory B-cell pool, down regulate EBV gene expression and persist in a latent state. Viral latency can be a long-term event and the association of EBV with an infected individual is considered to be for life. EBV is associated with the development of several types of cancer associated with lymphocytes or epithelial cells, principally Burkitt’s lymphoma, Hodgkin’s disease, and nasopharyngeal carcinoma. Primary infection with EBV can also result in infectious mononucleosis.
Prof Sinclair's research group investigate the interactions between EBV and human cells. After infection of human cells the virus establishes viral latency and promotes cancer development. When the infected cells differentiate the virus becomes reactivated and it undergoes the viral lytic replication cycle - producing hundreds of copies of infectious virus.
Epigenetic control of Epstein–Barr virus transcription – relevance to viral life cycle?
DNA methylation normally leads to silencing of gene expression but Epstein–Barr virus (EBV) provides an exception to the epigenetic paradigm.
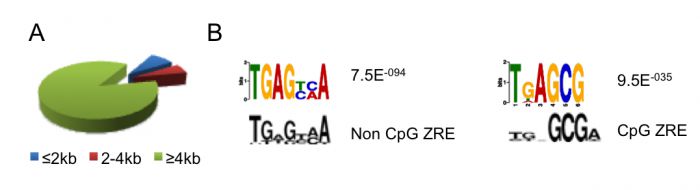
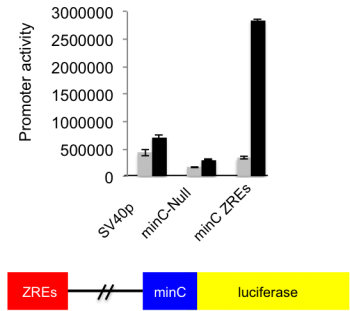
DNA methylation is absolutely required for the expression of many viral genes. Although the viral genome is initially un-methylated in newly infected cells, it becomes extensively methylated during the establishment of viral latency. One of the major regulators of EBV gene expression is a viral transcription factor called Zta (BZLF1, ZEBRA, Z) that resembles the cellular AP1 transcription factor. Zta recognizes at least 32 variants of a 7-nucleotide DNA sequence element, the Zta-response element (ZRE), some of which contain a CpG motif. Zta only binds to the latter class of ZREs in their DNA-methylated form, whether they occur in viral or cellular promoters and is functionally relevant for the activity of these promoters. The ability of Zta to interpret the differential DNA methylation of the viral genome is paramount for both the establishment of viral latency and the release from latency to initiate viral replication.
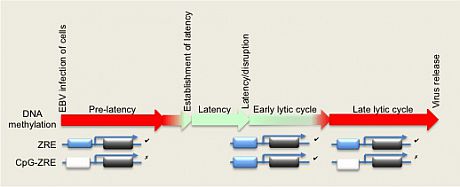 In the figure, the colored bar depicts the methylation state of EBV genome during different phases of the viral life cycle, with non-methylated DNA in red and methylated DNA represented in green. Two types of Zta-responsive gene are shown below: those containing ZREs that are independent of DNA methylation (ZRE) and those that are dependent on methylation (CpG-ZRE). Note that non-methylated CpG-ZREs cannot be bound by Zta.
In the figure, the colored bar depicts the methylation state of EBV genome during different phases of the viral life cycle, with non-methylated DNA in red and methylated DNA represented in green. Two types of Zta-responsive gene are shown below: those containing ZREs that are independent of DNA methylation (ZRE) and those that are dependent on methylation (CpG-ZRE). Note that non-methylated CpG-ZREs cannot be bound by Zta.
The DNA methyation status of the EBV genome dictates whether the ZREs are recognized by Zta. Those that can be recognized are shown in blue and those that can not be are shown in white.The CpG-ZREs are only recognized during latency and early lytic cycle.
See review on this subject
Sinclair Frontiers in Genetics 2012
Methylation-dependent DNA binding

The protein that is the master regulator of EBV replication, Zta, interacts directly with DNA. Zta interacts with specific short DNA sequence elements in the regulatory regions of genes and in the origin of replication. Some sequence elements contain a CpG dinucleotide and are only recognised when the CpG dinucleotide is methylated.
Zta is the prototype for a new family of methylation dependent transcription factors which include the cellular transcription factor C/EBP alpha.
By the interaction with methylated DNA sequence elements, Zta is able to overturn epigenetic silencing of both host and viral gene expression. This has profound implications for the establishment of viral latency, the reactivation of virus from latency and the reprogramming of host gene expression.
We developed computational tools to identify the location of the DNA binding elements that Zta interacts with in collaboration with Dr Sue Jones's research group. These have been used to mine the human and viral genomes to identify genes potentially regulated by Zta. This revealed that EBV overturns epigenetic silencing of both viral and host genes.
see our research publications
Karlsson, PLOS pathogens 2008;
Karlsson et al Biochem Soc transactions 2008;
Heather et al Journal of General Virology 2009;
Flower et al PLOS One 2010; Flower et al PLOS One 2011 and
Ramasubramanyan et al 2012
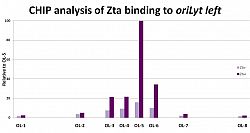
Zta binding sites on the viral genome identified using Chromatin-precipitation-linked to DNA sequencing.
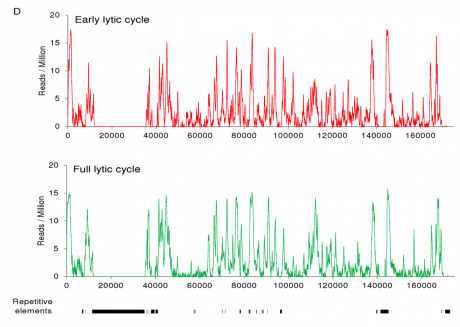
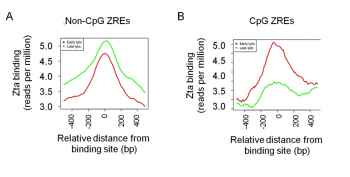
We identified Zta binding sites on the viral genome using ChIP-DNA sequencing. This revealed that Zta interacts with the majority of promoters in the viral genome and has a sustained inpact on gene exrpssion throught the EBV lytic cycle. Furthermore it revealed on a genome-wide scale that interaction with CpG containing ZREs is dpendent on the epigenetic mark of DNA methylation.
see our research publication
Ramasubramanyan et al 2012
DNA damage response pathway and EBV replication
Using a global proteomic approach, we identified a host DNA damage repair protein that specifically interacts with Zta: 53BP1.
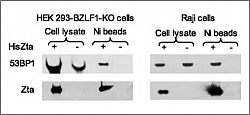 53BP1 is intimately connected with the ATM signal transduction pathway, which is activated during EBV replication. The interaction of 53BP1 with Zta requires the C-terminal ends of both proteins. A series of Zta mutants that show a wild-type ability to perform basic functions of Zta, such as dimer formation, interaction with DNA, and the transactivation of viral genes, were shown to have lost the ability to induce the viral lytic cycle. Each of these mutants also is compromised in the C-terminal region for interaction with 53BP1. In addition, the knockdown of 53BP1 expression reduced viral replication, suggesting that the association between Zta and 53BP1 is involved in the viral replication cycle.
53BP1 is intimately connected with the ATM signal transduction pathway, which is activated during EBV replication. The interaction of 53BP1 with Zta requires the C-terminal ends of both proteins. A series of Zta mutants that show a wild-type ability to perform basic functions of Zta, such as dimer formation, interaction with DNA, and the transactivation of viral genes, were shown to have lost the ability to induce the viral lytic cycle. Each of these mutants also is compromised in the C-terminal region for interaction with 53BP1. In addition, the knockdown of 53BP1 expression reduced viral replication, suggesting that the association between Zta and 53BP1 is involved in the viral replication cycle.
See our research publications
Bailey et al Journal of Virology 2009
Ramasubramanyan et al 2012
Chromatin conformation of EBV genome in latency and during latency disruption
The chromatin environment around the silent lytic genes in EBV genome is complex during latency, containing both activation and repression elements.
Our data show that H3K9me3 and H3K27me3 are enriched on the EBV genome. Both are associated with silenced regions of cell genomes through polycomb-mediated gene repression and heterochromatin mediated gene repression. We hypothesize that both contribute to silencing the EBV promoters required for lytic replication cycle during latency and this contributes to the maintenance of viral latency.
See our research publication
Ramasubramanyan et al 2012
We identified host genes regulated during viral replication using genome-wide RNA sequencing
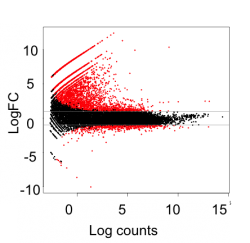
This identified 2263 cellular genes that are regulated during EBV lytic replicaiton cycle.
See our research publication
Ramasubramanyan et al 2015
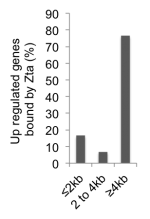
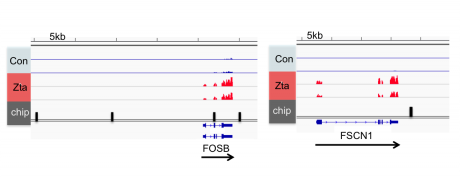
We identified Zta binding sites on the human genome using Chromatin-precipitation-linked to DNA sequencing.

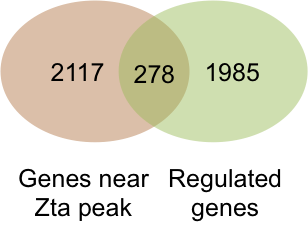
We integrated the transcriptome and the binding sitedata ets and identified the cellular genes that are directly regulated by Zta during lytic cycle
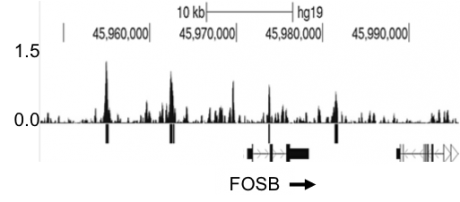
This research showed that Zta binds at enhancer elements far away from transcriptional start sites.
See our research publication
Ramasubramanyan et al 2015