Prof Michelle West- Epstein-Barr virus-driven transcriptional reprogramming in carcinogenesis
The West group studies the molecular mechanisms of transcriptional regulation by the key transcription factors expressed by the cancer-associated Epstein-Barr virus in cells infected and immortalised by the virus. The group is working to decipher the transcriptional changes involved in B cell carcinogenesis with a focus on gene activation and silencing through epigenetic changes established through the exploitation of long-range regulatory elements (enhancers) by viral factors. The group are also using biophysics and structural biology to determine how EBV transcription factors hijack cellular factors to control gene expression.
Prof Alison Sinclair -Transcriptional control during cancer virus replication
The Sinclair group investigate how transcription of viral and host cell genes are controlled in cells infected with the cancer-associated Epstein-Barr virus (EBV) when the virus initiates the replicative phase of its life cycle to produce hundreds of copies of infectious virus. Their work focuses on an EBV transcription factor Zta that is critical for lytic replication. Current research focuses on the mechanisms of transcriptional regulation of the viral and cellular genomes and the relevance of the cellular gene regulation for viral replication in Burkitt’s Lymphoma and Nasopharyngeal carcinoma.
Dr Mark Paget-Transcriptional control in bacteria
The Paget group studies the molecular biology of gene expression in bacteria with focus on the actinomycete family of bacteria which includes many bacteria of medical and industrial importance. The Streptomyces genus are the source of most clinically useful antibiotics and the major human pathogen Mycobacterium tuberculosis kills more people world-wide than any other single infectious agent. The group use the primary model organism Streptomyces coelicolor to study transcriptional control mechanisms in response to environmental signals such as nutrient limitation, oxygen limitation and oxidative stress and are also investigating gene regulation in the control of fermentation in the bioethanol-producing thermophile Geobacillus thermoglucosidasius.
Dr Sarah Newbury-RNA stability in development and microRNAs as biomarkers
Research in the Newbury group focuses on the role of RNA degradation in animal development. Degradation of messenger RNAs and microRNAs is controlled by ribonucleases and other factors that work together as a “molecular machine”. The group are studying the ways in which this novel mechanism of gene regulation can control stem cell differentiation, proliferation and apoptosis in the model organism Drosophila. More recently, the group has started work on elucidation of the role of one of these ribonucleases in the progression of the bone cancer osteosarcoma. In collaboration with clinical researchers, the group are also identifying microRNAs as diagnostic and prognostic biomarkers for human diseases including myeloma, melanoma and sepsis.
Prof Simon Morley-mRNA utilisation and translation in eukaryotes
Work in the Morley group investigates the signalling pathways regulating mRNA utilisation in eukaryotic cells during proliferation and differentiation. The main focus of the group is on the initiation factor complex, eIF4F, and its regulated assembly during different phases of the cell cycle. They are also developing tools to investigate localised protein synthesis in cells maintained in 2D and 3D culture. Although the regulation of protein synthesis is fundamental to cell growth and survival, relatively little is actually known about the role of phosphorylation of translation initiation factors in modulating this process and this is a key focus of their research.
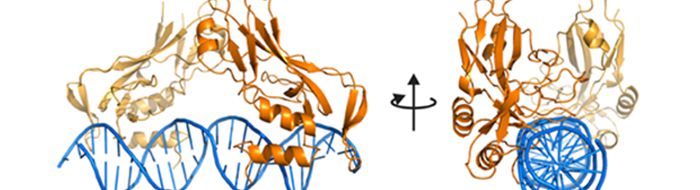
Dr Erika Mancini-Structural biology of chromatin remodelling and transcriptional regulation
Work in the Mancini group aims to determine the structural basis underlying the interplay between chromatin remodelling factors, transcription factors and DNA, a crucial requirement for the precise regulation of eukaryotic gene expression. The group are studying the molecular details of how transcription factors and chromatin remodelling ATPases work, alone or in complex, to define an active or repressed gene transcription state using X-ray crystallography in combination with Cryo-EM, NMR, SAXS and a range of biochemical and biophysical techniques.
Prof Juan-Pablo Couso-The identification and characterization of new genes encoding small peptides
Work in the Couso lab studies novel genes with small Open Reading Frames (smORFs). Their work has contributed to the creation of a new field that has the potential to change the way we perceive what a gene is, and to expand our current view of the information content of a genome. The group are currently investigating the translation of smORFs during development and tissue function and the function of smORFs at the genomic, cellular and molecular levels, including their role in key gene regulation pathways such as Notch and Wnt.
Dr Claudio Alonso- RNA regulation during neural development
Work in the Alonso group investigates the molecular mechanisms controlling gene function during animal development. The group studies the function and regulation of a specific group of genes, the Hox genes, which encode an evolutionary conserved family of transcriptional regulators required for the correct head-to-tail patterning of animal bodies. We are currently using our close understanding of the Hox system to study: (i) the molecular mechanisms underlying alternative splicing, alternative polyadenylation, mRNA degradation and microRNA regulation in vivo, and (ii) the ways these RNA regulatory processes are linked to specific biological functions during neural development.